 |
CASToR
1.0
Tomographic Reconstruction (PET/SPECT)
|
 |
CASToR
1.0
Tomographic Reconstruction (PET/SPECT)
|
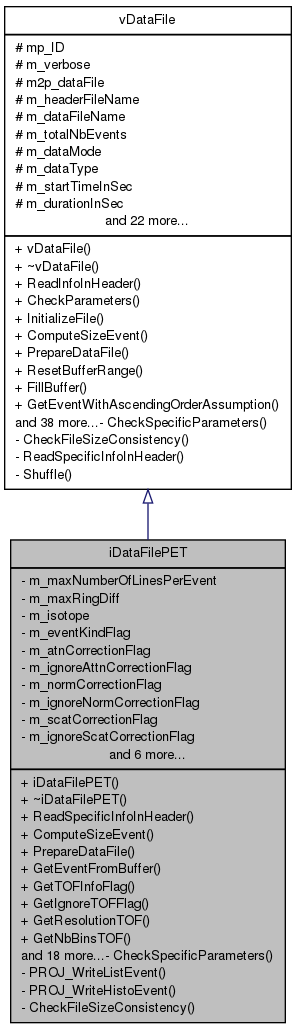
Inherit from vDataFile. Class that manages the reading of a PET input file (header + data). More...
#include <iDataFilePET.hh>

Public Member Functions | |
| iDataFilePET () | |
| iDataFilePET constructor. Initialize the member variables to their default values. | |
| ~iDataFilePET () | |
| iDataFilePET destructor. | |
| int | ReadSpecificInfoInHeader () |
| Read through the header file and gather specific PET information. | |
| int | ComputeSizeEvent () |
| Computation of the size of each event according to the mandatory/optional correction fields. | |
| int | PrepareDataFile () |
| Store different kind of information inside arrays (data relative to specific correction as well as basic raw data for the case data is loaded in RAM) Use the flag provided by the user to determine how the data has to be sorted (preloaded or read on the fly) | |
| vEvent * | GetEventFromBuffer (char *ap_buffer, int a_th) |
| Read an event from the position pointed by 'ap_buffer', parse the generic or modality-specific information, and store them in the (multithreaded) 'm2p_BufferEvent' object. | |
| bool | GetTOFInfoFlag () |
| bool | GetIgnoreTOFFlag () |
| FLTNB | GetResolutionTOF () |
| int | GetNbBinsTOF () |
| FLTNB | GetBinSizeTOF () |
| int | GetMaxRingDiff () |
| void | SetMaxNberLinesPerEvent (uint16_t a_value) |
| set the max number of line per event in the datafile | |
| void | SetIsotope (string a_value) |
| initialize the isotope string value | |
| void | SetTOFResolution (FLTNB a_value) |
| initialize the TOF resolution (for list-mode) | |
| void | SetNbTOFBins (int a_value) |
| initialize the number of TOF bins (for histogram-mode) | |
| void | SetIgnoreTOFFlag (bool a_ignoreTOFFlag) |
| Set a boolean that that if we ignore TOF information or not. | |
| void | SetTOFBinSize (FLTNB a_value) |
| initialize the size of TOF bins (for histogram-mode) | |
| void | SetEventKindFlagOn () |
| set to true the flag indicating the presence of the kind of a list-mode event in the datafile TODO check if consistent with datafile type | |
| void | SetAtnCorrectionFlagOn () |
| set to true the flag indicating the presence of attenuation correction factors in the datafile | |
| void | SetNormCorrectionFlagOn () |
| set to true the flag indicating the presence of normalization correction factors in the datafile | |
| void | SetScatterCorrectionFlagOn () |
| set to true the flag indicating the presence of scatter correction factors in the datafile | |
| void | SetRandomCorrectionFlagOn () |
| set to true the flag indicating the presence of random correction factors in the datafile | |
| bool | GetNormCorrectionFlag () |
| Simply return m_normCorrectionFlag. | |
| bool | GetAtnCorrectionFlag () |
| Simply return m_atnCorrectionFlag. | |
| int | PROJ_InitFile () |
| Initialize the fstream objets for output writing as well as some other variables specific to the Projection script (Event-based correction flags, Estimated size of data file) | |
| int | PROJ_WriteEvent (vEvent *ap_Event, int a_th) |
| Write event according to the chosen type of data. | |
| int | PROJ_WriteHeader () |
| Generate a header file according to the projection and data output informations. Used by Projection algorithm. | |
| int | PROJ_GetScannerSpecificParameters () |
| Get PET specific parameters for projections from the scanner object, through the scannerManager. | |
Private Member Functions | |
| int | CheckSpecificParameters () |
| Check parameters specific to PET data. | |
| int | PROJ_WriteListEvent (iEventListPET *ap_Event, int a_th) |
| Write a PET list-mode event. | |
| int | PROJ_WriteHistoEvent (iEventHistoPET *ap_Event, int a_th) |
| Write a PET histogram event. | |
| int | CheckFileSizeConsistency () |
| This function is implemented in child classes Check if file size is consistent. | |
Private Attributes | |
| uint16_t | m_maxNumberOfLinesPerEvent |
| int | m_maxRingDiff |
| string | m_isotope |
| bool | m_eventKindFlag |
| bool | m_atnCorrectionFlag |
| bool | m_ignoreAttnCorrectionFlag |
| bool | m_normCorrectionFlag |
| bool | m_ignoreNormCorrectionFlag |
| bool | m_scatCorrectionFlag |
| bool | m_ignoreScatCorrectionFlag |
| bool | m_randCorrectionFlag |
| bool | m_ignoreRandCorrectionFlag |
| bool | m_TOFInfoFlag |
| bool | m_ignoreTOFFlag |
| FLTNB | m_resolutionTOF |
| int | m_nbBinsTOF |
| FLTNB | m_binSizeTOF |
Inherit from vDataFile. Class that manages the reading of a PET input file (header + data).
It contains several arrays corresponding to the different kind of informations the data file could contain.
As many booleans as arrays say if the data are here or not. The data file can be either completely loaded, or read event by event during reconstruction.
MPI is coming here to cut the data file into peaces (also either can be loaded or read on-the-fly).
Definition at line 26 of file iDataFilePET.hh.
iDataFilePET constructor. Initialize the member variables to their default values.
Definition at line 30 of file iDataFilePET.cc.
iDataFilePET destructor.
Definition at line 61 of file iDataFilePET.cc.
| int iDataFilePET::CheckFileSizeConsistency | ( | ) | [private, virtual] |
This function is implemented in child classes
Check if file size is consistent.
Implements vDataFile.
Definition at line 614 of file iDataFilePET.cc.
| int iDataFilePET::CheckSpecificParameters | ( | ) | [private, virtual] |
Check parameters specific to PET data.
Implements vDataFile.
Definition at line 534 of file iDataFilePET.cc.

| int iDataFilePET::ComputeSizeEvent | ( | ) | [virtual] |
Computation of the size of each event according to the mandatory/optional correction fields.
Implements vDataFile.
Definition at line 117 of file iDataFilePET.cc.

| iDataFilePET::GetAtnCorrectionFlag | ( | ) | [inline] |
Simply return m_atnCorrectionFlag.
Definition at line 207 of file iDataFilePET.hh.

| iDataFilePET::GetBinSizeTOF | ( | ) | [inline] |
Definition at line 105 of file iDataFilePET.hh.
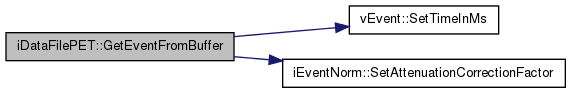
| vEvent * iDataFilePET::GetEventFromBuffer | ( | char * | ap_buffer, |
| int | a_th | ||
| ) | [virtual] |
Read an event from the position pointed by 'ap_buffer', parse the generic or modality-specific information, and store them in the (multithreaded) 'm2p_BufferEvent' object.
| ap_buffer | : address pointing to the event to recover |
| a_th | : index of the thread from which the function was called |
Implements vDataFile.
Definition at line 321 of file iDataFilePET.cc.
| iDataFilePET::GetIgnoreTOFFlag | ( | ) | [inline] |
Definition at line 87 of file iDataFilePET.hh.

| iDataFilePET::GetMaxRingDiff | ( | ) | [inline, virtual] |
Reimplemented from vDataFile.
Definition at line 111 of file iDataFilePET.hh.
| iDataFilePET::GetNbBinsTOF | ( | ) | [inline] |
Definition at line 99 of file iDataFilePET.hh.

| iDataFilePET::GetNormCorrectionFlag | ( | ) | [inline] |
Simply return m_normCorrectionFlag.
Definition at line 200 of file iDataFilePET.hh.

| iDataFilePET::GetResolutionTOF | ( | ) | [inline] |
Definition at line 93 of file iDataFilePET.hh.

| iDataFilePET::GetTOFInfoFlag | ( | ) | [inline] |
Definition at line 81 of file iDataFilePET.hh.

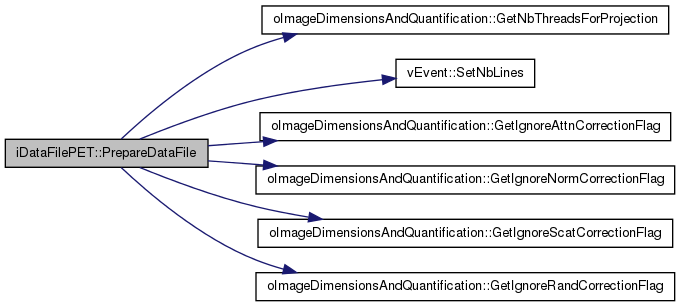
| int iDataFilePET::PrepareDataFile | ( | ) | [virtual] |
Store different kind of information inside arrays (data relative to specific correction as well as basic raw data for the case data is loaded in RAM)
Use the flag provided by the user to determine how the data has to be sorted (preloaded or read on the fly)
Implements vDataFile.
Definition at line 201 of file iDataFilePET.cc.

| int iDataFilePET::PROJ_GetScannerSpecificParameters | ( | ) | [virtual] |
Get PET specific parameters for projections from the scanner object, through the scannerManager.
Implements vDataFile.
Definition at line 1201 of file iDataFilePET.cc.

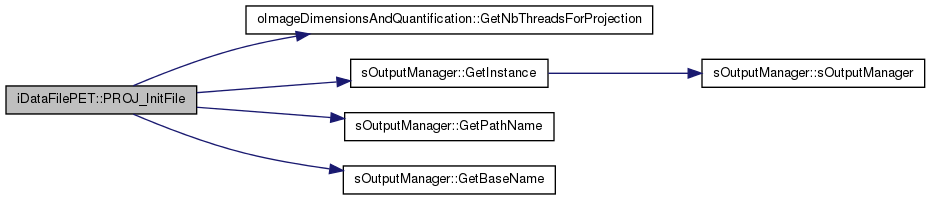
| int iDataFilePET::PROJ_InitFile | ( | ) | [virtual] |
Initialize the fstream objets for output writing as well as some other variables specific to the Projection script (Event-based correction flags, Estimated size of data file)
Implements vDataFile.
Definition at line 768 of file iDataFilePET.cc.

| int iDataFilePET::PROJ_WriteEvent | ( | vEvent * | ap_Event, |
| int | a_th | ||
| ) | [virtual] |
Write event according to the chosen type of data.
| ap_Event | : event containing the data to write |
| a_th | : index of the thread from which the function was called |
Implements vDataFile.
Definition at line 836 of file iDataFilePET.cc.

| int iDataFilePET::PROJ_WriteHeader | ( | ) | [virtual] |
Generate a header file according to the projection and data output informations.
Used by Projection algorithm.
Implements vDataFile.
Definition at line 1066 of file iDataFilePET.cc.

| int iDataFilePET::PROJ_WriteHistoEvent | ( | iEventHistoPET * | ap_Event, |
| int | a_th | ||
| ) | [private] |
Write a PET histogram event.
| ap_Event | : event containing the data to write |
| a_th | : index of the thread from which the function was called |
Definition at line 875 of file iDataFilePET.cc.

| int iDataFilePET::PROJ_WriteListEvent | ( | iEventListPET * | ap_Event, |
| int | a_th | ||
| ) | [private] |
Write a PET list-mode event.
| ap_Event | : event containing the data to write |
| a_th | : index of the thread from which the function was called |
Definition at line 955 of file iDataFilePET.cc.

| int iDataFilePET::ReadSpecificInfoInHeader | ( | ) | [virtual] |
Read through the header file and gather specific PET information.
Implements vDataFile.
Definition at line 74 of file iDataFilePET.cc.
| void iDataFilePET::SetAtnCorrectionFlagOn | ( | ) | [inline] |
set to true the flag indicating the presence of attenuation correction factors in the datafile
This function is dedicated to datafile conversion scripts
Definition at line 172 of file iDataFilePET.hh.

| iDataFilePET::SetEventKindFlagOn | ( | ) | [inline] |
set to true the flag indicating the presence of the kind of a list-mode event in the datafile TODO check if consistent with datafile type
This function is dedicated to datafile conversion scripts
Definition at line 165 of file iDataFilePET.hh.
| iDataFilePET::SetIgnoreTOFFlag | ( | bool | a_ignoreTOFFlag | ) | [inline] |
Set a boolean that that if we ignore TOF information or not.
| a_flag |
Definition at line 149 of file iDataFilePET.hh.
| iDataFilePET::SetIsotope | ( | string | a_value | ) | [inline] |
initialize the isotope string value
| a_value |
The name should corresponds to one corresponding tag in the isotope configuration file in config/. This function is dedicated to datafile conversion scripts
Definition at line 126 of file iDataFilePET.hh.

| iDataFilePET::SetMaxNberLinesPerEvent | ( | uint16_t | a_value | ) | [inline] |
set the max number of line per event in the datafile
Definition at line 117 of file iDataFilePET.hh.

| iDataFilePET::SetNbTOFBins | ( | int | a_value | ) | [inline] |
initialize the number of TOF bins (for histogram-mode)
| a_value |
This function is dedicated to datafile conversion scripts
Definition at line 142 of file iDataFilePET.hh.
| iDataFilePET::SetNormCorrectionFlagOn | ( | ) | [inline] |
set to true the flag indicating the presence of normalization correction factors in the datafile
This function is dedicated to datafile conversion scripts
Definition at line 179 of file iDataFilePET.hh.

| iDataFilePET::SetRandomCorrectionFlagOn | ( | ) | [inline] |
set to true the flag indicating the presence of random correction factors in the datafile
This function is dedicated to datafile conversion scripts
Definition at line 193 of file iDataFilePET.hh.

| iDataFilePET::SetScatterCorrectionFlagOn | ( | ) | [inline] |
set to true the flag indicating the presence of scatter correction factors in the datafile
This function is dedicated to datafile conversion scripts
Definition at line 186 of file iDataFilePET.hh.

| iDataFilePET::SetTOFBinSize | ( | FLTNB | a_value | ) | [inline] |
initialize the size of TOF bins (for histogram-mode)
| a_value |
This function is dedicated to datafile conversion scripts
Definition at line 157 of file iDataFilePET.hh.
| iDataFilePET::SetTOFResolution | ( | FLTNB | a_value | ) | [inline] |
initialize the TOF resolution (for list-mode)
| a_value |
This function is dedicated to datafile conversion scripts
Definition at line 134 of file iDataFilePET.hh.
bool iDataFilePET::m_atnCorrectionFlag [private] |
Flag that says if attenuation correction terms are included in the data. Default = false
Definition at line 292 of file iDataFilePET.hh.
FLTNB iDataFilePET::m_binSizeTOF [private] |
Size of TOF bins for histogram mode. Default = -1.0
Definition at line 304 of file iDataFilePET.hh.
bool iDataFilePET::m_eventKindFlag [private] |
Flag for informations about the event nature (true, scatter, random) in the data. Default = false
Definition at line 291 of file iDataFilePET.hh.
bool iDataFilePET::m_ignoreAttnCorrectionFlag [private] |
Flag to say if we ignore the attenuation correction even if present. Default = false
Definition at line 293 of file iDataFilePET.hh.
bool iDataFilePET::m_ignoreNormCorrectionFlag [private] |
Flag to say if we ignore the normalization correction even if present. Default = false
Definition at line 295 of file iDataFilePET.hh.
bool iDataFilePET::m_ignoreRandCorrectionFlag [private] |
Flag to say if we ignore the random correction even if present. Default = false
Definition at line 299 of file iDataFilePET.hh.
bool iDataFilePET::m_ignoreScatCorrectionFlag [private] |
Flag to say if we ignore the scatter correction even if present. Default = false
Definition at line 297 of file iDataFilePET.hh.
bool iDataFilePET::m_ignoreTOFFlag [private] |
Flag to say if we ignore the TOF data even if present, or not. Default = false
Definition at line 301 of file iDataFilePET.hh.
string iDataFilePET::m_isotope [private] |
Isotope. Default = unknown
Definition at line 290 of file iDataFilePET.hh.
uint16_t iDataFilePET::m_maxNumberOfLinesPerEvent [private] |
Number of lines in each event in the datafile. Default = 1
Definition at line 288 of file iDataFilePET.hh.
int iDataFilePET::m_maxRingDiff [private] |
Max ring difference between 2 crystals in a LOR. Default value calculated from the scanner files
Definition at line 289 of file iDataFilePET.hh.
int iDataFilePET::m_nbBinsTOF [private] |
Number of TOF bins for histogram mode. Default = 1
Definition at line 303 of file iDataFilePET.hh.
bool iDataFilePET::m_normCorrectionFlag [private] |
Flag that says if normalization correction terms are included in the data. Default = false
Definition at line 294 of file iDataFilePET.hh.
bool iDataFilePET::m_randCorrectionFlag [private] |
Flag that says if random correction terms are included in the data. Default = false
Definition at line 298 of file iDataFilePET.hh.
FLTNB iDataFilePET::m_resolutionTOF [private] |
TOF resolution in mm. Default = -1.0
Definition at line 302 of file iDataFilePET.hh.
bool iDataFilePET::m_scatCorrectionFlag [private] |
Flag that says if scatter correction terms are included in the data. Default = false
Definition at line 296 of file iDataFilePET.hh.
bool iDataFilePET::m_TOFInfoFlag [private] |
Flag that says if TOF information is included in the data. Default = false
Definition at line 300 of file iDataFilePET.hh.
 1.7.6.1
1.7.6.1